Evaluation¶
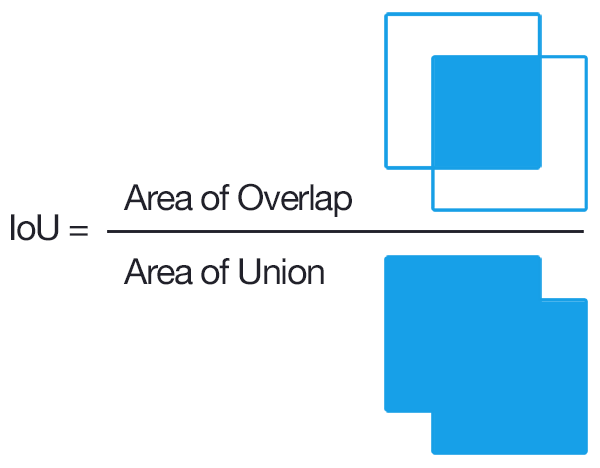
To convert overlap among predicted and ground truth bounding boxes into measures of accuracy and precision, the most common approach is to compare the overlap using the intersection-over-union metric (IoU). IoU is the ratio between the area of the overlap between the predicted polygon box and the ground truth polygon box divided by and the area of the combined bounding box region.
Let’s start by getting some sample data and predictions
from deepforest import evaluate
from deepforest import main
from deepforest import get_data
from deepforest import visualize
import os
import pandas as pd
m = main.deepforest()
m.use_release()
csv_file = get_data("OSBS_029.csv")
predictions = m.predict_file(csv_file=csv_file, root_dir=os.path.dirname(csv_file))
predictions.head()
xmin ymin xmax ymax label score image_path
0 330.0 342.0 373.0 391.0 Tree 0.802979 OSBS_029.tif
1 216.0 206.0 248.0 242.0 Tree 0.778803 OSBS_029.tif
2 325.0 44.0 363.0 82.0 Tree 0.751573 OSBS_029.tif
3 261.0 238.0 296.0 276.0 Tree 0.748605 OSBS_029.tif
4 173.0 0.0 229.0 33.0 Tree 0.738209 OSBS_029.tif
ground_truth = pd.read_csv(csv_file)
ground_truth.head()
image_path xmin ymin xmax ymax label
0 OSBS_029.tif 203 67 227 90 Tree
1 OSBS_029.tif 256 99 288 140 Tree
2 OSBS_029.tif 166 253 225 304 Tree
3 OSBS_029.tif 365 2 400 27 Tree
4 OSBS_029.tif 312 13 349 47 Tree
visualize.plot_prediction_dataframe(predictions, ground_truth, root_dir = os.path.dirname(csv_file))
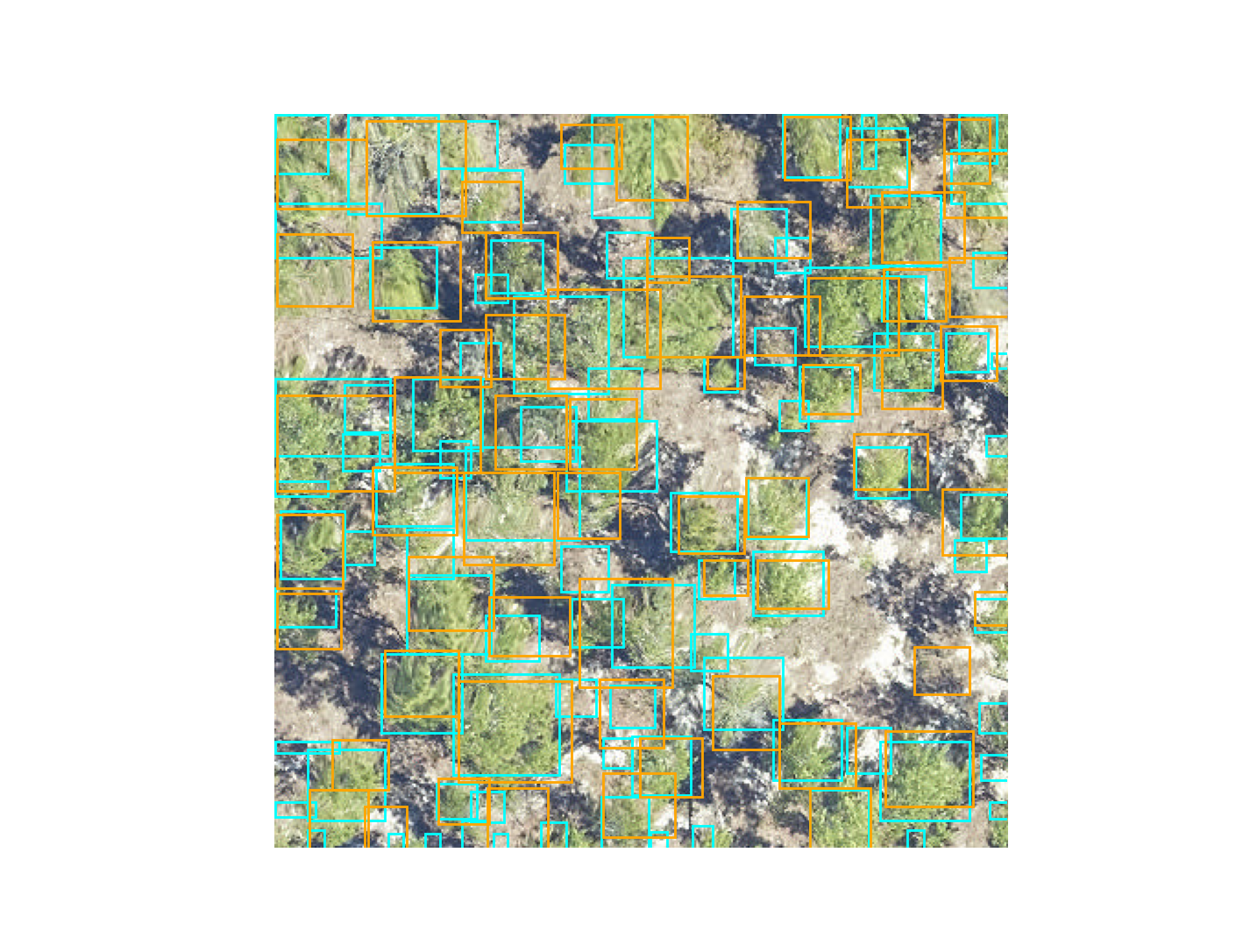
The IoU metric ranges between 0 (no overlap) to 1 (perfect overlap). In the wider computer vision literature, the conventional threshold value for overlap is 0.5, but this value is arbitrary and does not ultimately relate to any particular ecological question. We considered boxes which have an IoU score of greater than 0.4 as true positive, and scores less than 0.4 as false negatives. The 0.4 value was chosen based on visual evaluation of the threshold that indicated a good visual match between the predicted and observed crown. We tested a range of overlap thresholds from 0.3 (less overlap among matching crowns) to 0.6 (more overlap among matching crowns) and found that 0.4 balanced a rigorous cutoff without spuriously removing trees that would be useful for downstream analysis.
After computing the IoU for the ground truth data, we get the resulting dataframe.
result = evaluate.evaluate_image(predictions=predictions, ground_df=ground_truth, show_plot=True, root_dir=os.path.dirname(csv_file), savedir=None)
result.head()
prediction_id truth_id IoU predicted_label true_label
90 90 0 0.059406 Tree Tree
65 65 1 0.335366 Tree Tree
17 17 2 0.578551 Tree Tree
50 50 3 0.532902 Tree Tree
34 34 4 0.595862 Tree Tree
Where prediction_id is a unique ID to each predicted tree box. truth is a unique ID to each ground truth box. The predicted and true labels are tree in this, case but could generalize to multi-class problems. From here we can calculate precision and recall at a given IoU metric.
result["match"] = result.IoU > 0.4
true_positive = sum(result["match"])
recall = true_positive / result.shape[0]
precision = true_positive / predictions.shape[0]
recall
0.819672131147541
precision
0.5494505494505495
This can be stated as 81.97% of the ground truth boxes are correctly matched to a predicted box at IoU threshold of 0.4, and 54.94% of predicted boxes match a ground truth box. Optimally we want a model that is both precise and accurate.
The above logic is wrapped into the evaluate.evaluate() function
result = evaluate.evaluate(predictions=predictions, ground_df=ground_truth,root_dir=os.path.dirname(csv_file), savedir=None)
This is a dictionary with keys
result.keys()
dict_keys(['results', 'box_precision', 'box_recall', 'class_recall'])
The added class_recall dataframe is mostly relevant for multi-class problems, in which the recall and precision per class is given.
result["class_recall"]
label recall precision size
0 Tree 1.0 0.67033 61
One important decision was how to average precision and recall across multiple images. Two reasonable options might be to take all predictions and all ground truth and compute the statistic on the entire dataset. This strategy makes more sense for evaluation data that are relatively homogenous across images. We prefer to take the average of per-image precision and recall. This helps balanace the dataset if some images have many trees, and other have few trees, such as when you are comparing multiple habitat types. Users are welcome to calculate their own statistics directly from the results dataframe.
result["results"].head()
prediction_id truth_id IoU ... true_label image_path match
90 90 0 0.059406 ... Tree OSBS_029.tif False
65 65 1 0.335366 ... Tree OSBS_029.tif False
17 17 2 0.578551 ... Tree OSBS_029.tif True
50 50 3 0.532902 ... Tree OSBS_029.tif True
34 34 4 0.595862 ... Tree OSBS_029.tif True